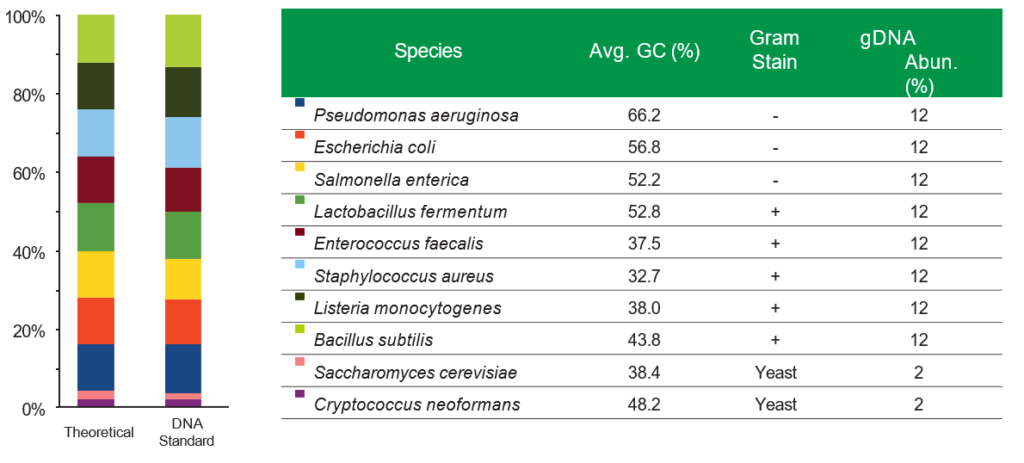
DNA from three Gram-negative bacteria, five Gram-positive bacteria, and two tough-to-lyse yeasts. The ZymoBIOMICS® Microbial Community Standards are perfect for assessing bias in popular extraction methods. The microbial standards are accurately characterized, with a wide GC range (15% – 85%) and contain negligible impurities (< 0.01%), enabling easy exposure of artifacts, errors, and bias in microbiomics or metagenomic workflows.
| Product | Cat. No. | Size |
|---|---|---|
| ZymoBIOMICS® Microbial Community Standard | D6300 | 10 preps |
| ZymoBIOMICS® Microbial Community DNA Standard | D6306 | 2,000 ng |
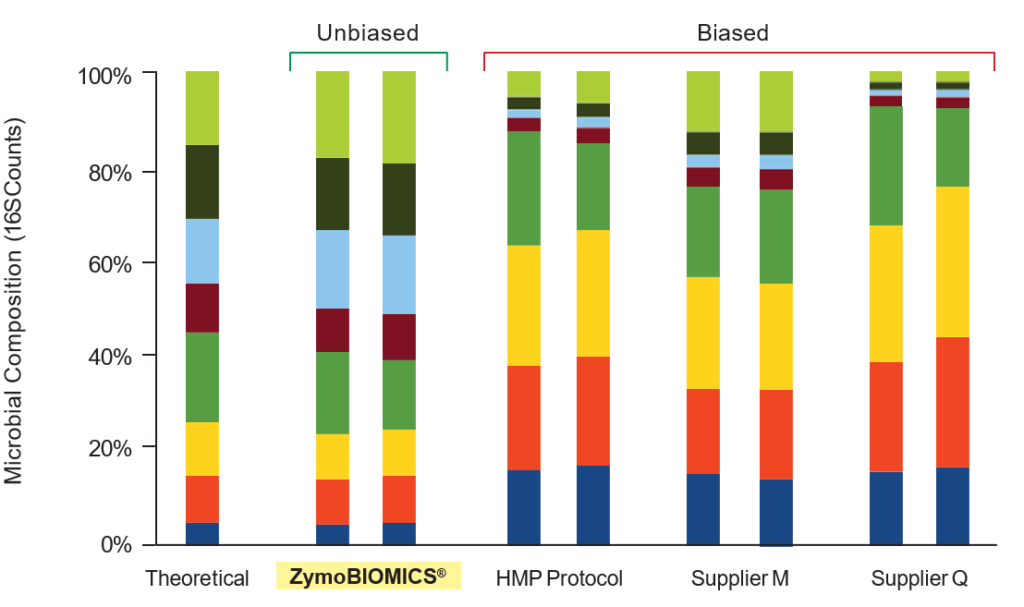
The ZymoBIOMICS® DNA Miniprep Kit provides accurate representation of the organisms extracted from the ZymoBIOMICS® Microbial Community Standard.
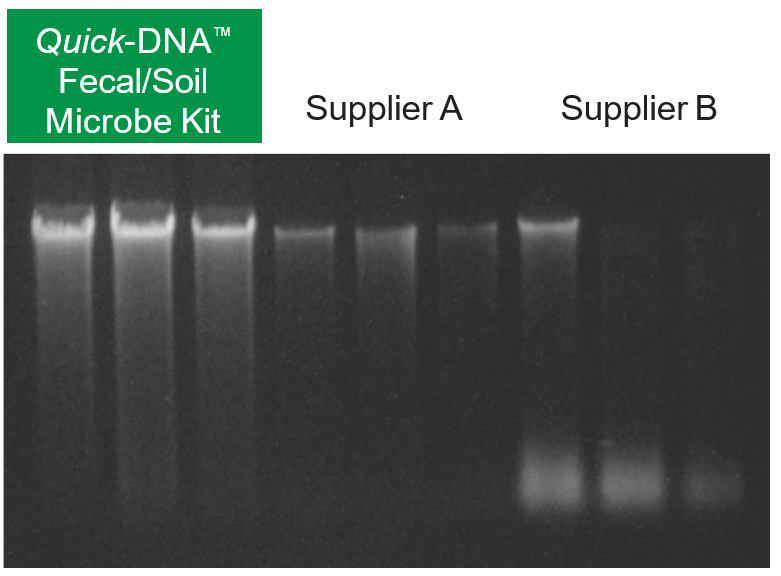
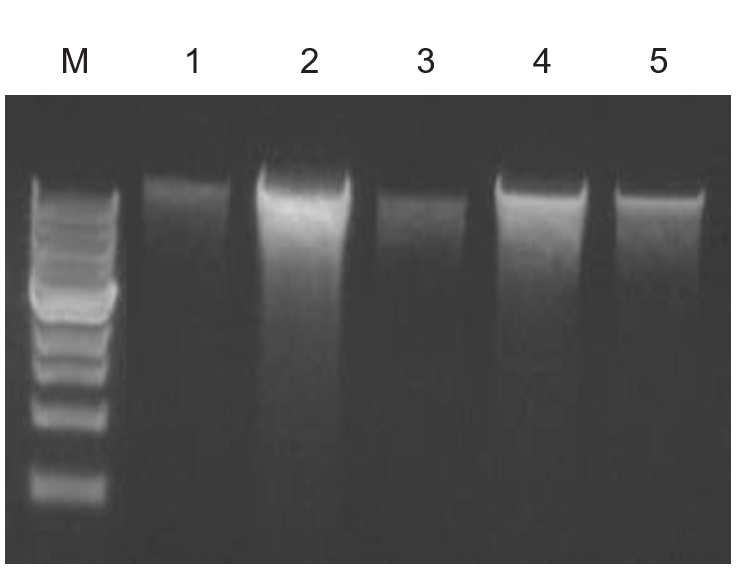
High-quality total DNA was isolated from different environmental sample sources using the Quick-DNA™ Fecal/Soil Microbe Kit and compared against other suppliers. (A) Equivalent amounts of feces were processed using each kit, then equal volumes of eluted DNA were analyzed on a 0.8% (w/v) agarose gel stained with EtBr. (B) Metagenomic DNA isolated from 5 soil samples. M: 1 kb marker (NEB); 1-5: soil samples (sand, sandy clay loam, hydrophobic sandy loam course, sandy loam, fine gravel).
| Product | Cat. No. | Size | |
|---|---|---|---|
| ZymoBIOMICS® DNA Miniprep Kit | D4300 | 50 preps | T3007S (4 tubes) |
| Quick-DNA™ Fecal/Soil Microbe Miniprep Kit | D6010 | 50 preps | - |
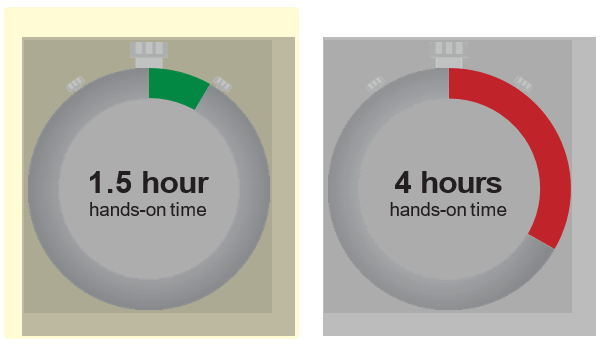
The Quick-16S™ NGS Library Prep Kit is >2.5 times faster than the conventional 16S library prep method. The Quick-16S™ Kit simplifies the 16S library prep workflow by quantifying libraries using qPCR, instead of TapeStation® analyses, and by using a single-tube library cleanup
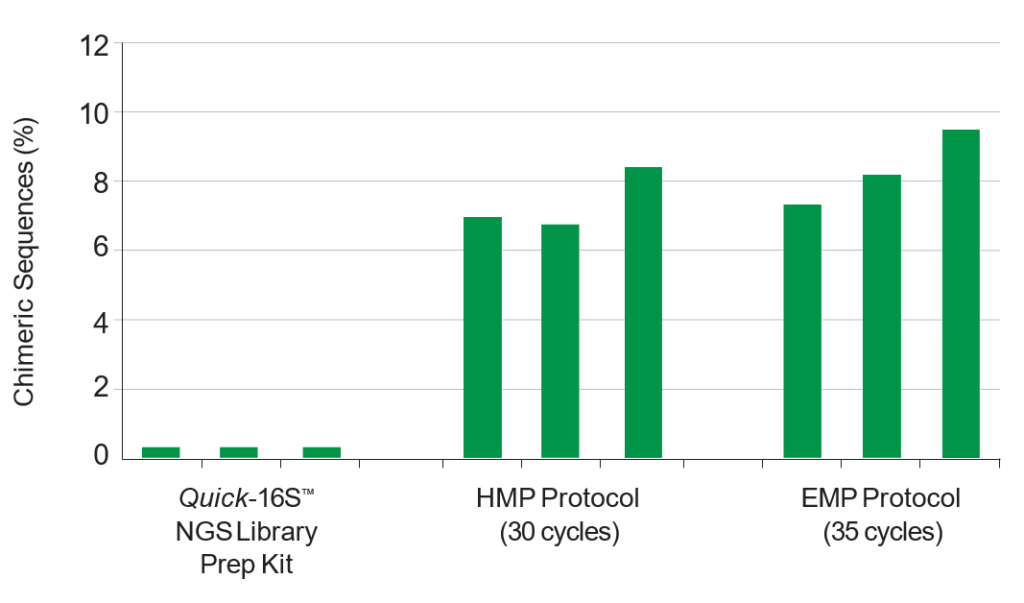
The Quick-16S™ NGS Library Prep Kit minimizes PCR chimera formation compared to two common protocols: Human Microbiome Project (HMP) and Earth Microbiome Project (EMP). Equivalent amounts of the same fecal DNA sample were used as input. Chimeric sequences were predicted with Uchime (https://www.drive5.com/uchime).
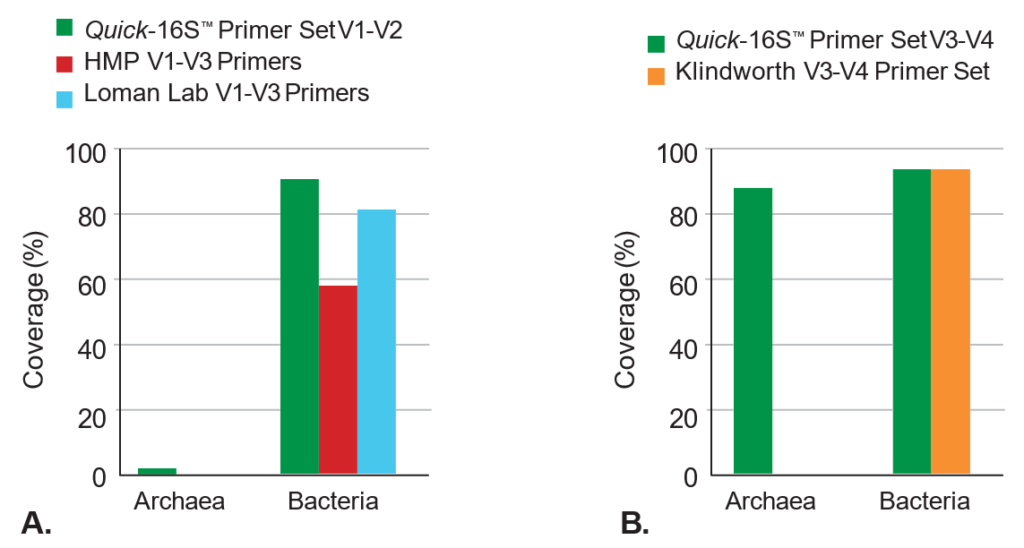
A.The Quick-16S™ Primer Set V1-V2 includes coverage of common human- associated microbes, including Bifidobacterium, Propionibacterium, and Chlamydia, which are missed in common V1-V2 or V1-V3 primers.
B.The Quick-16S™ Primer Set V3-V4 provides up to 87% coverage for archaea, organisms commonly found in the human gut. The common V3-V4 primers provide 0% coverage for archaea.
Unlocking Microbiome Insight
8h-17h Thứ Hai đến Thứ Sáu
Tầng 4, Tòa nhà PLS, 457-459A Nguyễn Đình Chiểu, Phường Bàn Cờ, Thành phố Hồ Chí Minh
info@pacificinformatics.com.vn
+84 28 3719 8877
Bản quyền © 2025 Pacinfo. Đã đăng ký bản quyền.